不同領域的數據科學家使用聚類方法在他們的數據集中找到自然的“相似”觀察組。流行的聚類方法可以是:
基于質心的:根據與某個質心的接近程度將點分組為 k 組。
基于圖形的:根據圖中頂點的連接對其進行分組。
Density-based:根據附近區域數據的密度或稀疏性更靈活地分組。
基于層次密度的應用程序空間聚類 w / Noise (HDBSCAN)算法是一種density-based聚類方法,對噪聲具有魯棒性(將稀疏區域中的點作為簇邊界,并將其中一些點直接標記為噪聲)。基于密度的聚類方法,如 HDBSCAN ,能夠發現形狀奇特、大小各異的聚類 — 與k-means、k-medioids或高斯混合模型等基于質心的聚類方法截然不同,這些方法找到一組 k 個質心,將簇建模為固定形狀和大小的球。除了必須預先指定 k 之外,基于質心的算法的性能和簡單性幫助它們仍然是高維聚類點的最流行方法之一;即使在不修改輸入數據點的情況下,它們也無法對不同大小、形狀或密度的簇進行建模。
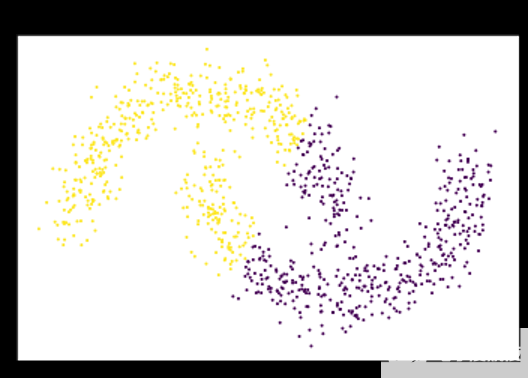
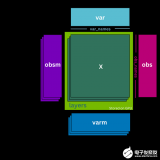
圖 1 : K-means 假設數據可以用固定大小的高斯球建模,并切割衛星,而不是單獨聚類。 K-means 將每個點指定給一個簇,即使存在噪聲和異常值也會影響結果的質心s.
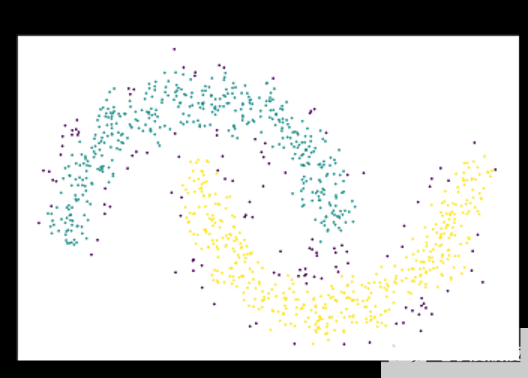
圖 2 :基于密度的算法通過從密集區域的突出鄰域擴展簇來解決此問題。 DBSCAN 和 HDBSCAN 可以將這些點解釋為噪聲,并將其標記為噪聲,如圖中的紫色點。
HDBSCAN 建立在一種眾所周知的基于密度的聚類算法 DBSCAN 的基礎上,該算法不要求提前知道簇的數量,但仍然存在一個不幸的缺點,即假設簇可以用一個全局密度閾值建模。這使得對具有不同密度的簇進行建模變得困難。 HDBSCAN 改進了這一缺點,通過使用單鏈聚簇來構建樹狀圖,從而可以找到不同密度的簇。另一種著名的基于密度的聚類方法稱為光學算法,它改進了 DBSCAN ,并使用分層聚類來發現密度不同的聚類。光學技術通過將點投影到一個新的空間(稱為可達空間)來改進標準的單鏈接聚類,該空間將噪聲從密集區域進一步移開,使其更易于處理。然而,與許多其他層次聚集聚類方法(如單鏈接聚類和完全鏈接聚類)一樣, OPTICS 也存在以單個全局切割值切割生成的樹狀圖的缺點。 HDBSCAN 本質上是光學+ DBSCAN ,引入了集群穩定性的度量,以在不同級別上切割樹狀圖。
我們將通過快速示例演示 HDBSCAN 的 RAPIDS cuML 實現中當前支持的功能,并將提供我們在 GPU 上實現的一些實際示例和基準。在閱讀了這篇博文之后,我們希望您對 RAPIDS ‘ GPU – 加速 HDBSCAN 實施可以為您的工作流和探索性數據分析過程帶來的好處感到興奮。
RAPIDS 中的 HDBSCAN 入門
GPU 提供了一組 RAPIDS – 加速 CPU 庫,幾乎可以替代 PyData 生態系統中許多流行的庫。下面的示例筆記本演示了 Python 上使用最廣泛的 HDBSCAN Python 庫與 GPU 上的 RAPIDS cuML HDBSCAN 之間的 API 兼容性(擾流板警報–在許多情況下,它與更改導入一樣簡單)。
Basic Usage
Example of training an HDBSCAN model using the hdbscan Python package in Scikit-learn contrib:
from sklearn import datasets
from hdbscan import HDBSCAN
X, y = datasets.make_moons(n_samples=50, noise=0.05)
model = HDBSCAN(min_samples=5)
y_hat = model.fit_predict(X)
from sklearn import datasets
from cuml.cluster import HDBSCAN
X, y = datasets.make_moons(n_samples=50, noise=0.05)
model = HDBSCAN(min_samples=5, gen_min_span_tree=True)
y_hat = model.fit_predict(X)
/share/workspace/cuml/python/cuml/internals/api_decorators.py:794: FutureWarning: Pass handle=None, verbose=False, output_type=None as keyword args. From version 21.06, passing these as positional arguments will result in an error return func(**kwargs)
Plotting
We can plot the minimum spanning tree the same way we would for the original HDBSCAN implementation:
model.minimum_spanning_tree_.plot()
The single linkage dendrogram and condensed tree can be plotted as well:
model.single_linkage_tree_.plot()
model.condensed_tree_.plot()
下面是一個非常簡單的示例,演示了基于密度的聚類優于基于質心的技術對某些類型數據的好處,以及使用 HDBSCAN 優于 DBSCAN 的好處。
Clustering Algorithm Comparison
Example notebook showing the strengths of density-based clustering techniques DBSCAN & HDBSCAN on datasets with odd and interleaved shapes.
Generate the data
from sklearn import datasets
import numpy as np
import warnings
warnings.filterwarnings("ignore")
X, y = datasets.make_moons(n_samples=1000, noise=0.12)
import matplotlib.pyplot as plt
plt.scatter(X[:,0], X[:,1], c=y, s=0.5)
K-Means
Even when we know there are only 2 clusters in this dataset, we can see that k-means just separates the space into two evenly-sized regions. It's not possible to model the interleaved and odd cluster shapes without performing some heavy pre-processing to project the points into a space where they can be modeled with fixed-sized balls.
from cuml.cluster import KMeans
kmeans_labels_ = KMeans(n_clusters=2).fit_predict(X)
plt.title("Interleaved Moons w/ K-Means")
plt.xticks([])
plt.yticks([])
plt.scatter(X[:,0], X[:,1], c=kmeans_labels_, s=1.0)
DBSCAN
While this particular example dataset tends to be a great demonstration of how DBSCAN can find clusters of odd shapes and similar densities, theepsparameter sets the radius of a ball around each point where points inside the ball are considered part of its neighborhood. This neighborhood is called an epsilon neighborhoods and it can be pretty non-intuitive to tune in practice. In addition, it's assumed that the sameepssetting applies globally to all the data points. A little bit more intuitive is themin_samplesparameter, which determines the neighborhood size within theepsball for a point to be considered acore point, upon which its neighborhood will be expanded to form a cluster.
from cuml.cluster import DBSCAN
As we can see, the default paramter settings of DBSCAN (and other density-based algorithms) make assumptions about the data that won't often lead to good clustering results.
dbscan_labels_ = DBSCAN().fit_predict(X)
plt.scatter(X[:,0], X[:,1], c=dbscan_labels_, s=1.0)
We can set themin_samplesparameter to 10, but the amount of noise is too high for this alone to result in good cluster assignments.
dbscan_labels_ = DBSCAN(min_samples=10).fit_predict(X)
import numpy as np
np.unique(dbscan_labels_)
array([0], dtype=int32)
plt.scatter(X[:,0], X[:,1], c=dbscan_labels_, s=0.5)
We can introduce a setting for theepsparameter, but it's unclear how to determine a good setting for this value just from this visualization. Setting this value too small can yield too many small clusterings which are not representative of the larger patterns in the data.
dbscan_labels_ = DBSCAN(min_samples=10, eps=0.08).fit_predict(X)
np.unique(dbscan_labels_)
array([-1, 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20], dtype=int32)
Increasingepsto 0.12 did the trick and we notice it found the points which surround the dense regions as noise (these will have a label of -1)
dbscan_labels_ = DBSCAN(min_samples=10, eps=0.12).fit_predict(X)
plt.title("Interleaved Moons w/ DBSCAN")
plt.xticks([])
plt.yticks([])
plt.scatter(X[:,0], X[:,1], c=dbscan_labels_, s=0.5)
HDBSCAN
HDBSCAN removes the need to specify a global epsilon neighborhood radius around each point (eps) and instead uses themin_pointsargument to create a radius to its k-th nearest neighbor, using the radius to push sparser regions further away from each other. Since HDBSCAN is built upon the agglomerative clustering technique single-linkage clustering, each point starts as its own cluster and is merged into a tree, bottom-up, until a single root cluster is reached. Amin_cluster_sizeargument allows us to specify a minimum threshold for when clusters in the agglomerated tree should be merged. The dendrogram is cut at varying levels, or selected, using a measure of cluster stability, based on the distances between the points in each cluster and the clusters from which they originated. Clusters which are selected cosume all of their descendent clusters. HDBSCAN provides an additional optioncluster_selection_epsilonto set the minimum distance threshold from which clusters will be split up into smaller clusters.
from cuml.cluster import HDBSCAN
labels_ = HDBSCAN(min_samples=10).fit_predict(X)
np.unique(labels_)
array([-1, 0, 1], dtype=int32)
plt.scatter(X[:,0], X[:,1], c=labels_, s=0.5)
Adding the same clustering against the reference Python HDBSCAN implementation
from hdbscan import HDBSCAN as HDBSCANref
labels_ = HDBSCANref(min_samples=25).fit_predict(X)
plt.scatter(X[:,0], X[:,1], c=labels_, s=0.5)
HDBSCAN 在實踐中的應用
基于密度的聚類技術自然適合于許多不同的聚類任務,因為它們能夠找到形狀奇特、大小各異的聚類。與許多其他通用機器學習算法一樣,沒有免費的午餐,因此盡管 HDBSCAN 改進了一些成熟的算法,但它仍然不是完成這項工作的最佳工具。盡管如此, DBSCAN 和 HDBSCAN 在從地理空間和協同過濾/推薦系統到金融和科學計算等領域的應用中取得了顯著的成功,被應用于從天文學到加速器物理學到基因組學等學科。它對噪聲的魯棒性也使得它對于異常值和異常檢測應用非常有用。
與數據分析和機器學習生態系統中的許多其他工具一樣,計算時間對生產系統和迭代工作流有很大的影響。更快的 HDBSCAN 意味著能夠嘗試更多的想法并制作更好的模型。下面是幾個使用 HDBSCAN 對單詞嵌入和單細胞 RNA 基因表達進行聚類的示例筆記本。這些都是為了簡短,并為您自己的數據集使用 HDBSCAN 提供了一個很好的起點。您是否已成功地將 HDBSCAN 應用于工業或科學領域,我們在此未列出?請留下評論,因為我們很想聽到。如果您在自己的硬件上運行示例筆記本電腦,還請告知我們您的設置以及您使用 RAPIDS 的經驗。
單詞嵌入
向量嵌入代表了一種流行且非常廣泛的聚類機器學習應用。我們之所以選擇 GoogleNews 數據集,是因為它足夠大,可以很好地顯示我們的算法的規模,但又足夠小,可以在一臺機器上執行。下面的筆記本演示了如何使用 HDBSCAN 查找有意義的主題,這些主題來自單詞嵌入的角度空間中的高密度區域,并使用 UMAP 可視化生成的主題簇。它使用整個數據集的一個子集進行可視化,但為調整不同的超參數和熟悉它們對結果集群的影響提供了一個很好的演示。我們使用默認的超參數設置(形狀為 3Mx300 )對整個數據集進行了基準測試,并在 24 小時后停止了 CPU 上的 Scikit learn contrib 實現。 RAPIDS 的實現大約需要 22 .8 分鐘。
Clustering and Visualizing Embeddings with HDBSCAN & UMAP
In this notebook, we use a dataset of word embeddings to extract areas that could be associated with meaningful topics. We use HDBSCAN to find the meaningful topics since it can account for noise when labeling regions of high density without explicitly requiring a distance as input. We also use UMAP to visualize the resulting topic clusters.
This notebook requires theGoogleNews Word2Vec dataset, which can be downloaded fromhttps://drive.google.com/file/d/0B7XkCwpI5KDYNlNUTTlSS21pQmM/edit?usp=sharing.
from gensim import models
import cupy as cp
from cuml.manifold import UMAP
from cuml.preprocessing import normalize
from cuml.cluster import HDBSCAN
import matplotlib.pyplot as plt
from matplotlib.pyplot import figure
w = models.KeyedVectors.load_word2vec_format("GoogleNews-vectors-negative300.bin", binary=True)
We can usemin_samplesto control the radius of the core distances. A smaller setting leads to more clusters and less points being treated as noise.min_cluster_sizeandmax_cluster_sizebound the number of clusters by only allowing points to form a cluster when they are >min_cluster_clusterand splitting a single cluster into multiple smaller clusters when it is >max_cluster_size
n_points = 100000
umap_n_neighbors=100
umap_min_dist=1e-3
umap_spread=2.0
umap_n_epochs=500
umap_random_state=42
hdbscan_min_samples=25
hdbscan_min_cluster_size=5
hdbscan_max_cluster_size=1000
hdbscan_cluster_selection_method="leaf"
L2 normalize the vectors so that the Euclidean distance between them will give the angular distance.
%%time
X = normalize(cp.array(w.vectors))[:n_points]
CPU times: user 4.17 s, sys: 4.57 s, total: 8.73 s Wall time: 8.76 s
X.shape
(100000, 300)
First, we can use manifold learning to convert the neighborhoods of points in the angular distance space to a 2-dimensional Euclidean space so that we can better virualize the clusters.
%%time
umap = UMAP(n_neighbors=umap_n_neighbors,
min_dist=umap_min_dist,
spread=umap_spread,
n_epochs=umap_n_epochs,
random_state=umap_random_state)
embeddings = umap.fit_transform(X)
CPU times: user 1.71 s, sys: 910 ms, total: 2.62 s Wall time: 2.62 s
%%time
hdbscan = HDBSCAN(min_samples=hdbscan_min_samples,
min_cluster_size=hdbscan_min_cluster_size,
max_cluster_size=hdbscan_max_cluster_size,
cluster_selection_method=hdbscan_cluster_selection_method)
labels = hdbscan.fit_predict(X)
Label prop iterations: 23 Label prop iterations: 10 Label prop iterations: 6 Label prop iterations: 5 Iterations: 4 17663,114,1077,12,218,1603 CPU times: user 5.6 s, sys: 933 ms, total: 6.53 s Wall time: 6.51 s
Percentage of points were labeled as meaningful topics
len(labels[labels!=-1]) / embeddings.shape[0]
0.04558
How many labels are there?
len(cp.unique(labels))
151
x = embeddings[:,0]
y = embeddings[:,1]
x = x[labels>-1]
y = y[labels>-1]
labels_nonoise = labels[labels>-1]
x_noise = embeddings[:,0]
y_noise = embeddings[:,1]
x_noise = x_noise[labels==-1]
y_noise = y_noise[labels==-1]
We observe that the regions of high density where points are collected closely together have filled out nicely into clusters of various different shapes and sizes. We interpret the distances of points within their local neighborhoods in the following plot as being more similar than points in disconnected neighborhoods. It's not uncommon for UMAP to split neighborhoods that into multiple clusters even though HDBSCAN labeled them as a single cluster. This just implies that the neighborhoods of the points which are split are more similar to each other internally than they are across their multiple separated components.
figure(figsize=(10, 7), dpi=80)
plt.scatter(x.get(), y.get(), c=labels_nonoise.get(), s=0.1, cmap='gist_ncar')
We can also visualize the noise
figure(figsize=(10, 7), dpi=80)
plt.scatter(x_noise.get(), y_noise.get(), s=0.001, cmap="gray", alpha=0.4)
label_dist = cp.histogram(labels, bins=cp.unique(labels))
plt.plot(label_dist[1][:len(label_dist[0])-1].get(), label_dist[0][1:].get())
[]
Let's look at a few groups of terms that clustered together
for c in range(0, 10):
print("Cluster: %s, %s" % (c, [w.index_to_key[i] for i in cp.argwhere(labels==c)[:10,0].get()]))
Cluster: 0, ['##th', 'fourth', 'fifth', 'sixth', 'seventh', 'eighth', 'ninth', '###th', '##st', '##nd'] Cluster: 1, ['miles', 'north', 'south', 'west', 'east', 'kilometers', 'km', 'northwest', 'northeast', 'southwest'] Cluster: 2, ['petitioner', 'Defendant', 'Plaintiff', 'Appellant', 'Petitioner', 'appellants', 'Id.'] Cluster: 3, ['memorial', 'cemetery', 'burial', 'graves', 'remembrance'] Cluster: 4, ['FPL', 'AEP', 'Progress_Energy', 'TXU', 'FirstEnergy', 'Ameren', 'ComEd', 'NIPSCO'] Cluster: 5, ['Officials', 'Students', 'Residents', 'Customers', 'Businesses', 'Activists'] Cluster: 6, ['gay', 'gay_marriage', 'gays', 'lesbian', 'homosexual', 'LGBT', 'lesbians', 'homosexuals'] Cluster: 7, ['borrowings', 'Common_Stock', 'Common_Shares', 'Senior_Notes', 'senior_unsecured', 'debentures', 'convertible_notes', 'Preferred_Stock', 'Debentures', 'convertible_debentures'] Cluster: 8, ['-#.#', '-1', '-2', '-3', '-4'] Cluster: 9, ['Supreme_Court', 'justices', 'Alito', 'appellate_court', 'Samuel_Alito']
We can also order them according to their probabilities (notice most probabilities are fairly large because the regions are already pretty dense)
for c in range(0, 25):
indices = list(cp.argwhere(labels==c).get()[:,0])
sorted(indices, key=lambda x: 1 - hdbscan.probabilities_[x])
print("Cluster: %s, %s" % (c, [w.index_to_key[i] for i in indices[:10]]))
Cluster: 0, ['##th', 'fourth', 'fifth', 'sixth', 'seventh', 'eighth', 'ninth', '###th', '##st', '##nd'] Cluster: 1, ['miles', 'north', 'south', 'west', 'east', 'kilometers', 'km', 'northwest', 'northeast', 'southwest'] Cluster: 2, ['petitioner', 'Defendant', 'Plaintiff', 'Appellant', 'Petitioner', 'appellants', 'Id.'] Cluster: 3, ['memorial', 'cemetery', 'burial', 'graves', 'remembrance'] Cluster: 4, ['FPL', 'AEP', 'Progress_Energy', 'TXU', 'FirstEnergy', 'Ameren', 'ComEd', 'NIPSCO'] Cluster: 5, ['Officials', 'Students', 'Residents', 'Customers', 'Businesses', 'Activists'] Cluster: 6, ['gay', 'gay_marriage', 'gays', 'lesbian', 'homosexual', 'LGBT', 'lesbians', 'homosexuals'] Cluster: 7, ['borrowings', 'Common_Stock', 'Common_Shares', 'Senior_Notes', 'senior_unsecured', 'debentures', 'convertible_notes', 'Preferred_Stock', 'Debentures', 'convertible_debentures'] Cluster: 8, ['-#.#', '-1', '-2', '-3', '-4'] Cluster: 9, ['Supreme_Court', 'justices', 'Alito', 'appellate_court', 'Samuel_Alito'] Cluster: 10, ['Citigroup', 'Goldman_Sachs', 'Morgan_Stanley', 'UBS', 'Merrill_Lynch', 'Deutsche_Bank', 'HSBC', 'Credit_Suisse', 'JPMorgan', 'JP_Morgan'] Cluster: 11, ['CEO', 'chairman', 'vice_president', 'Director', 'Vice_President', 'managing_director', 'Executive_Director', 'Managing_Director', 'Senior_Vice', 'Executive_Vice'] Cluster: 12, ['Wal_Mart', 'Walmart', 'Kmart', 'Kroger', 'Costco', 'Nordstrom', 'JC_Penney'] Cluster: 13, ['drilling', 'mineralization', 'intersected', 'g_t_Au', 'drill_holes', 'g_t_gold', 'g_t', 'mineralized', 'molybdenum', 'gold_mineralization'] Cluster: 14, ['JB', 'JM', 'ML', 'JS', 'JL', 'JH'] Cluster: 15, ['Warner_Bros.', 'Lionsgate', 'DreamWorks', 'Paramount_Pictures', 'Sony_Pictures'] Cluster: 16, ['artist', 'painting', 'paintings', 'painter', 'sculptures', 'artworks', 'watercolors', 'Paintings', 'canvases'] Cluster: 17, ['loans', 'mortgage', 'lenders', 'mortgages', 'borrowers', 'foreclosures', 'subprime', 'mortgage_lenders'] Cluster: 18, ['plane', 'aircraft', 'planes', 'airplane', 'jets', 'Airbus_A###', 'aircrafts'] Cluster: 19, ['apartments', 'condo', 'condominium', 'condos', 'Apartments', 'condominiums', 'townhouse', 'townhomes', 'Condominiums', 'townhome'] Cluster: 20, ['fell', 'jumped', 'climbed', 'slipped', 'risen', 'surged', 'plunged', 'slid', 'soared', 'dipped'] Cluster: 21, ['school', 'students', 'teachers', 'elementary', 'Elementary', 'kindergarten', 'preschool', 'elementary_schools', 'eighth_grade', 'eighth_graders'] Cluster: 22, ['professor', 'Professor', 'scientist', 'researcher', 'Ph.D.', 'associate_professor', 'master_degree', 'PhD', 'assistant_professor', 'bachelor_degree'] Cluster: 23, ['gun', 'firearm', 'rifle', 'handgun', 'pistol', 'pistols', 'assault_rifle', '.##_caliber', '.##_caliber_handgun', 'gauge_shotgun'] Cluster: 24, ['#.####', 'yen', 'greenback', 'EUR_USD', 'downtrend', 'USD_JPY', '#.####/##', 'GBP_USD', 'EURUSD', 'Japanese_Yen']
單細胞 RNA
下面是一個基于掃描和俱樂部庫中教程筆記本的工作流示例。本示例筆記本取自 RAPIDS 單單元示例存儲庫,其中還包含幾個筆記本,演示了 RAPIDS 用于單細胞和三級分析。在 DGX-1 (英特爾 40 核至強 CPU + NVIDIA V100 GPU )上,我們發現 HDBSCAN (在 GPU 上是~ 1s ,而不是具有多個 CPU 線程的~ 29s )使用了包含~ 70k 肺細胞基因表達的數據集上的前 50 個主成分,加速了 29x 。
RAPIDS & Scanpy Single-Cell RNA-seq Workflow
Copyright (c) 2020, NVIDIA CORPORATION.
Licensed under the Apache License, Version 2.0 (the "License") you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
This notebook demonstrates a single-cell RNA analysis workflow that begins with preprocessing a count matrix of size(n_gene, n_cell)and results in a visualization of the clustered cells for further analysis.
For demonstration purposes, we use a dataset of ~70,000 human lung cells from Travaglini et al. 2020 (https://www.biorxiv.org/content/10.1101/742320v2) and label cells using the ACE2 and TMPRSS2 genes. See the README for instructions to download this dataset.
Import requirements
import numpy as np
import scanpy as sc
import anndata
import time
import os
import cudf
import cupy as cp
from cuml.decomposition import PCA
from cuml.manifold import TSNE
from cuml.cluster import KMeans
import rapids_scanpy_funcs
import warnings
warnings.filterwarnings('ignore', 'Expected ')
warnings.simplefilter('ignore')
We use the RAPIDS memory manager on the GPU to control how memory is allocated.
import rmm
rmm.reinitialize(
managed_memory=True, # Allows oversubscription
pool_allocator=False, # default is False
devices=0, # GPU device IDs to register. By default registers only GPU 0.
)
cp.cuda.set_allocator(rmm.rmm_cupy_allocator)
Input data
In the cell below, we provide the path to the.h5adfile containing the count matrix to analyze. Please see the README for instructions on how to download the dataset we use here.
We recommend saving count matrices in the sparse .h5ad format as it is much faster to load than a dense CSV file. To run this notebook using your own dataset, please see the README for instructions to convert your own count matrix into this format. Then, replace the path in the cell below with the path to your generated.h5adfile.
input_file = "../data/krasnow_hlca_10x.sparse.h5ad"
if not os.path.exists(input_file):
print('Downloading import file...')
os.makedirs('../data', exist_ok=True)
wget.download('https://rapids-single-cell-examples.s3.us-east-2.amazonaws.com/krasnow_hlca_10x.sparse.h5ad',
input_file)
Set parameters
# marker genes
RIBO_GENE_PREFIX = "RPS" # Prefix for ribosomal genes to regress out
markers = ["ACE2", "TMPRSS2", "EPCAM"] # Marker genes for visualization
# filtering cells
min_genes_per_cell = 200 # Filter out cells with fewer genes than this expressed
max_genes_per_cell = 6000 # Filter out cells with more genes than this expressed
# filtering genes
n_top_genes = 5000 # Number of highly variable genes to retain
# PCA
n_components = 50 # Number of principal components to compute
# t-SNE
tsne_n_pcs = 20 # Number of principal components to use for t-SNE
# k-means
k = 35 # Number of clusters for k-means
# KNN
n_neighbors = 15 # Number of nearest neighbors for KNN graph
knn_n_pcs = 50 # Number of principal components to use for finding nearest neighbors
# UMAP
umap_min_dist = 0.3
umap_spread = 1.0
# Gene ranking
ranking_n_top_genes = 50 # Number of differential genes to compute for each cluster
start = time.time()
Load and Prepare Data
We load the sparse count matrix from anh5adfile using Scanpy. The sparse count matrix will then be placed on the GPU.
data_load_start = time.time()
%%time
adata = sc.read(input_file)
CPU times: user 125 ms, sys: 228 ms, total: 354 ms Wall time: 353 ms
adata.X.shape
(65662, 26485)
a = np.diff(adata.X.indptr)
a
array([1347, 1713, 1185, ..., 651, 1050, 2218], dtype=int32)
len(a[a<3000])
52985
We maintain the index of unique genes in our dataset:
%%time
genes = cudf.Series(adata.var_names)
sparse_gpu_array = cp.sparse.csr_matrix(adata.X)
CPU times: user 836 ms, sys: 586 ms, total: 1.42 s Wall time: 1.45 s
Verify the shape of the resulting sparse matrix:
sparse_gpu_array.shape
(65662, 26485)
And the number of non-zero values in the matrix:
sparse_gpu_array.nnz
126510394
data_load_time = time.time()
print("Total data load and format time: %s" % (data_load_time-data_load_start))
Total data load and format time: 1.8720729351043701
Preprocessing
preprocess_start = time.time()
Filter
We filter the count matrix to remove cells with an extreme number of genes expressed.
%%time
sparse_gpu_array = rapids_scanpy_funcs.filter_cells(sparse_gpu_array, min_genes=min_genes_per_cell, max_genes=max_genes_per_cell)
CPU times: user 535 ms, sys: 304 ms, total: 839 ms Wall time: 838 ms
Some genes will now have zero expression in all cells. We filter out such genes.
%%time
sparse_gpu_array, genes = rapids_scanpy_funcs.filter_genes(sparse_gpu_array, genes, min_cells=1)
CPU times: user 1.03 s, sys: 162 ms, total: 1.19 s Wall time: 1.19 s
The size of our count matrix is now reduced.
sparse_gpu_array.shape
(65462, 22058)
Normalize
We normalize the count matrix so that the total counts in each cell sum to 1e4.
%%time
sparse_gpu_array = rapids_scanpy_funcs.normalize_total(sparse_gpu_array, target_sum=1e4)
CPU times: user 415 μs, sys: 599 μs, total: 1.01 ms Wall time: 747 μs
Next, we log transform the count matrix.
%%time
sparse_gpu_array = sparse_gpu_array.log1p()
CPU times: user 42.7 ms, sys: 52.4 ms, total: 95.1 ms Wall time: 94.4 ms
Select Most Variable Genes
We will now select the most variable genes in the dataset. However, we first save the 'raw' expression values of the ACE2 and TMPRSS2 genes to use for labeling cells afterward. We will also store the expression of an epithelial marker gene (EPCAM).
%%time
tmp_norm = sparse_gpu_array.tocsc()
marker_genes_raw = {
("%s_raw" % marker): tmp_norm[:, genes[genes == marker].index[0]].todense().ravel()
for marker in markers
}
del tmp_norm
CPU times: user 208 ms, sys: 175 ms, total: 383 ms Wall time: 383 ms
Now, we convert the count matrix to an annData object.
%%time
adata = anndata.AnnData(sparse_gpu_array.get())
adata.var_names = genes.to_pandas()
CPU times: user 178 ms, sys: 52.9 ms, total: 231 ms Wall time: 229 ms
Using scanpy, we filter the count matrix to retain only the 5000 most variable genes.
%%time
sc.pp.highly_variable_genes(adata, n_top_genes=n_top_genes, flavor="cell_ranger")
adata = adata[:, adata.var.highly_variable]
CPU times: user 977 ms, sys: 26.5 ms, total: 1 s Wall time: 1 s
Regress out confounding factors (number of counts, ribosomal gene expression)
We can now perform regression on the count matrix to correct for confounding factors - for example purposes, we use the number of counts and the expression of ribosomal genes. Many workflows use the expression of mitochondrial genes (named starting withMT-).
ribo_genes = adata.var_names.str.startswith(RIBO_GENE_PREFIX)
We now calculate the total counts and the percentage of ribosomal counts for each cell.
%%time
n_counts = adata.X.sum(axis=1)
percent_ribo = (adata.X[:,ribo_genes].sum(axis=1) / n_counts).ravel()
n_counts = cp.array(n_counts).ravel()
percent_ribo = cp.array(percent_ribo).ravel()
CPU times: user 881 ms, sys: 32.3 ms, total: 914 ms Wall time: 913 ms
And perform regression:
%%time
sparse_gpu_array = cp.sparse.csc_matrix(adata.X)
CPU times: user 653 ms, sys: 114 ms, total: 767 ms Wall time: 766 ms
%%time
sparse_gpu_array = rapids_scanpy_funcs.regress_out(sparse_gpu_array, n_counts, percent_ribo)
CPU times: user 31.8 s, sys: 10.5 s, total: 42.3 s Wall time: 43.2 s
Scale
Finally, we scale the count matrix to obtain a z-score and apply a cutoff value of 10 standard deviations, obtaining the preprocessed count matrix.
%%time
sparse_gpu_array = rapids_scanpy_funcs.scale(sparse_gpu_array, max_value=10)
CPU times: user 119 ms, sys: 169 ms, total: 289 ms Wall time: 287 ms
preprocess_time = time.time()
print("Total Preprocessing time: %s" % (preprocess_time-preprocess_start))
Total Preprocessing time: 48.95587778091431
Cluster & Visualize
We store the preprocessed count matrix as an AnnData object, which is currently in host memory. We also add the expression levels of the marker genes as observations to the annData object.
%%time
genes = adata.var_names
adata = anndata.AnnData(sparse_gpu_array.get())
adata.var_names = genes
for name, data in marker_genes_raw.items():
adata.obs[name] = data.get()
CPU times: user 199 ms, sys: 92.1 ms, total: 292 ms Wall time: 291 ms
Reduce
We use PCA to reduce the dimensionality of the matrix to its top 50 principal components.
%%time
adata.obsm["X_pca"] = PCA(n_components=n_components, output_type="numpy").fit_transform(adata.X)
CPU times: user 1.14 s, sys: 1.41 s, total: 2.55 s Wall time: 2.55 s
UMAP + Density clustering
We can also visualize the cells using the UMAP algorithm in Rapids. Before UMAP, we need to construct a k-nearest neighbors graph in which each cell is connected to its nearest neighbors. This can be done conveniently using rapids functionality already integrated into Scanpy.
Note that Scanpy uses an approximation to the nearest neighbors on the CPU while the GPU version performs an exact search. While both methods are known to yield useful results, some differences in the resulting visualization and clusters can be observed.
%%time
sc.pp.neighbors(adata, n_neighbors=n_neighbors, n_pcs=knn_n_pcs, method='rapids')
CPU times: user 4.04 s, sys: 276 ms, total: 4.32 s Wall time: 4.3 s
The UMAP function from Rapids is also integrated into Scanpy.
%%time
sc.tl.umap(adata, min_dist=umap_min_dist, spread=umap_spread, method='rapids')
WARNING: .obsp["connectivities"] have not been computed using umap CPU times: user 246 ms, sys: 229 ms, total: 475 ms Wall time: 474 ms
Next, we use the Louvain algorithm for graph-based clustering, once again using therapidsoption in Scanpy.
%%time
import pandas as pd
from cuml.cluster import HDBSCAN
hdbscan = HDBSCAN(min_samples=5, min_cluster_size=30)
adata.obs['hdbscan_gpu'] = pd.Categorical(pd.Series(hdbscan.fit_predict(adata.obsm['X_pca'])))
CPU times: user 718 ms, sys: 465 ms, total: 1.18 s Wall time: 1.19 s
%%time
import pandas as pd
from hdbscan import HDBSCAN as refHDBSCAN
hdbscan = refHDBSCAN(min_samples=5, min_cluster_size=30, core_dist_n_jobs=-1)
adata.obs['hdbscan_cpu'] = pd.Categorical(pd.Series(hdbscan.fit_predict(adata.obsm['X_pca'])))
CPU times: user 23.5 s, sys: 1.2 s, total: 24.7 s Wall time: 29.6 s
We plot the cells using the UMAP visualization, and using the Louvain clusters as labels.
%%time
sc.pl.umap(adata, color=["hdbscan_gpu"])
CPU times: user 597 ms, sys: 12.2 ms, total: 609 ms Wall time: 607 ms
RAPIDS CUML 項目包括端到端 GPU 加速的 HDBSCAN ,并提供 Python 和 C ++ + API 。與 cuML 中的許多基于鄰域的算法一樣,它利用 Facebook 的費斯庫中的蠻力 kNN 來加速相互可達空間中 kNN 圖的構建。這是目前的一個主要瓶頸,我們正在研究通過精確和近似近鄰選項進一步改進它的方法。
CUML 還包括單連鎖層次聚類的實現,它提供了 C ++和 Python API 。 GPU – 加速單個鏈接算法需要計算最小生成樹的新原語。此原語基于圖形,因此可以在 cugraph 和 cuml 庫中重用。我們的實現允許重新啟動,這樣我們就可以連接一個斷開連接的 knn 圖,并通過不必在 GPU 內存中存儲整個成對距離矩陣來提高可伸縮性。
與大多數CUML算法中的C++一樣,這些依賴于我們的大多數基于ML和基于圖元的[VZX27 ]。最后,他們利用利蘭·麥克因內斯和約翰·希利所做的偉大工作到 GPU ——甚至加快了群集壓縮和選擇步驟,使數據盡可能多地保留在 GPU 上,并在數據規模擴展到數百萬時提供額外的性能提升。
基準
我們使用了 McInnes 等人在 CPU 上的參考實現提供的基準筆記本,將其與 cuML 的新 GPU 實現進行比較。參考實現針對低維情況進行了高度優化,我們將高維情況與大量使用 Facebook FAISS 庫的暴力實現進行了比較。
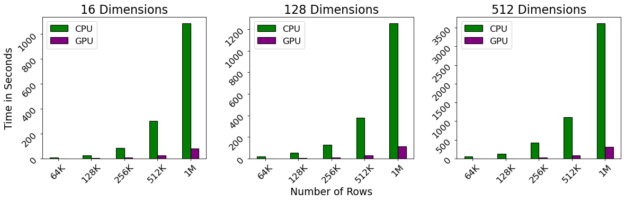
圖 4 : GPU – 加速 HDBSCAN 即使對于大型數據集也能保持近乎交互的性能,同時消除 CPU 有限的并行性。有了這些速度,你會發現你有更多的時間做其他事情,比如更好地了解你的數據。
基準測試是在 DGX-1 上執行的,該 DGX-1 包含 40 核 Intel 至強 CPU 和 NVIDIA 32gb V100 GPU s 。即使對維度數進行線性縮放,對行數進行二次縮放,我們觀察到 GPU 仍然保持接近交互性能,即使行數超過 1M 。
發生了什么變化?
雖然我們已經成功地在 GPU 上實現了 HDBSCAN 算法的核心,但仍有機會進一步提高其性能,例如通過加快蠻力 kNN 圖構造刪除距離計算,甚至使用近似 kNN。雖然歐幾里德距離涵蓋了最廣泛的用途,但我們還想公開Scikit 學習 Contrib 實現中提供的其他距離度量。
scikit learn contrib 實現還包含許多不錯的附加功能,這些功能沒有包含在 HDBSCAN 上的開創性論文中,例如半監督和模糊聚類。我們也有堅固的單連桿和光學算法的構建塊,這將是 RAPIDS 未來的良好補充。最后,我們希望在將來支持稀疏輸入。
如果您發現這些功能中的一個或多個可以使您的應用程序或數據分析項目更成功,即使此處未列出這些功能,請轉到我們的Github 項目并創建一個問題。
概括
HDBSCAN 是一種相對較新的基于密度的聚類算法“站在巨人的肩膀上”,改進了著名的 DBSCAN 和光學算法。事實上,它的核心原語還增加了重用,并為其他算法提供了構建塊,例如基于圖的最小生成樹和 RAPIDS ML 和圖庫中的單鏈接聚類。
與其他數據建模算法一樣, HDBSCAN 并不是所有工作的完美工具,但它在工業和科學計算應用中都有很多實際用途。它還可以與 PCA 或 UMAP 等降維算法配合使用,尤其是在探索性數據分析應用中。
關于作者
Corey Nolet 是 NVIDIA 的 RAPIDS ML 團隊的數據科學家兼高級工程師,他專注于構建和擴展機器學習算法,以支持光速下的極端數據負載。在 NVIDIA 工作之前, Corey 花了十多年時間為國防工業的 HPC 環境構建大規模探索性數據科學和實時分析平臺。科里持有英國理工學士學位計算機科學碩士學位。他還在攻讀博士學位。在同一學科中,主要研究圖形和機器學習交叉點的算法加速。科里熱衷于利用數據更好地了解世界。
Divye Gala 是 NVIDIA 的 RAPIDS 團隊的高級軟件工程師,在 GPU 上開發用于機器學習和圖形分析的快速可擴展算法。 Divye 持有女士和 Bs 。計算機科學學位。在空閑時間,迪維喜歡踢足球和看板球。
審核編輯:郭婷
-
cpu
+關注
關注
68文章
10825瀏覽量
211149 -
gpu
+關注
關注
28文章
4701瀏覽量
128705 -
API
+關注
關注
2文章
1485瀏覽量
61817
發布評論請先 登錄
相關推薦
GPU加速matlab程序
Nvidia宣布推出了一套新的開源RAPIDS庫
NVIDIA RAPIDS加速器可將工作分配集群中各節點
如何使用RAPIDS和CuPy時加速Gauss 秩變換
NVIDIA RAPIDS加速器v21.08的功能應用
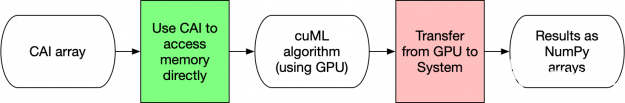
RAPIDS cuML中的輸入輸出可配置性
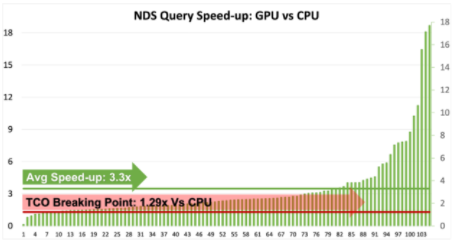
AutoML技術提高NVIDIA GPU和RAPIDS速度
利用Apache Spark和RAPIDS Apache加速Spark實踐
RAPIDS cuDF將pandas提速近150倍





 具有RAPIDS cuML的GPU加速分層DBSCAN
具有RAPIDS cuML的GPU加速分層DBSCAN










評論